Human genome at everybody's disposal - a review session
Another look at frameshift mutations
Indel gameToday we'll need
UCSC Genome Browser
(UC Santa Cruz Genome Browser) is a place on the Web where we can check where on the genome
(with respect to known genes and their internal structure) is a given variant located.
(An example of a variant lookup).
Note: to show only the canonical splice of a gene, see the instructions
here , the question "How can I show a single transcript per gene?"
Also check out the instructions on the Miro board
PDB database of protein structures.
A reminder from the last time: OMIM databse collects (for half a century now) the information about the connection between the gene variants and the human health.
GnomAd database collects the information about the variants in people without obvious disease phenotypes (though they might be carriers of recessive variants)
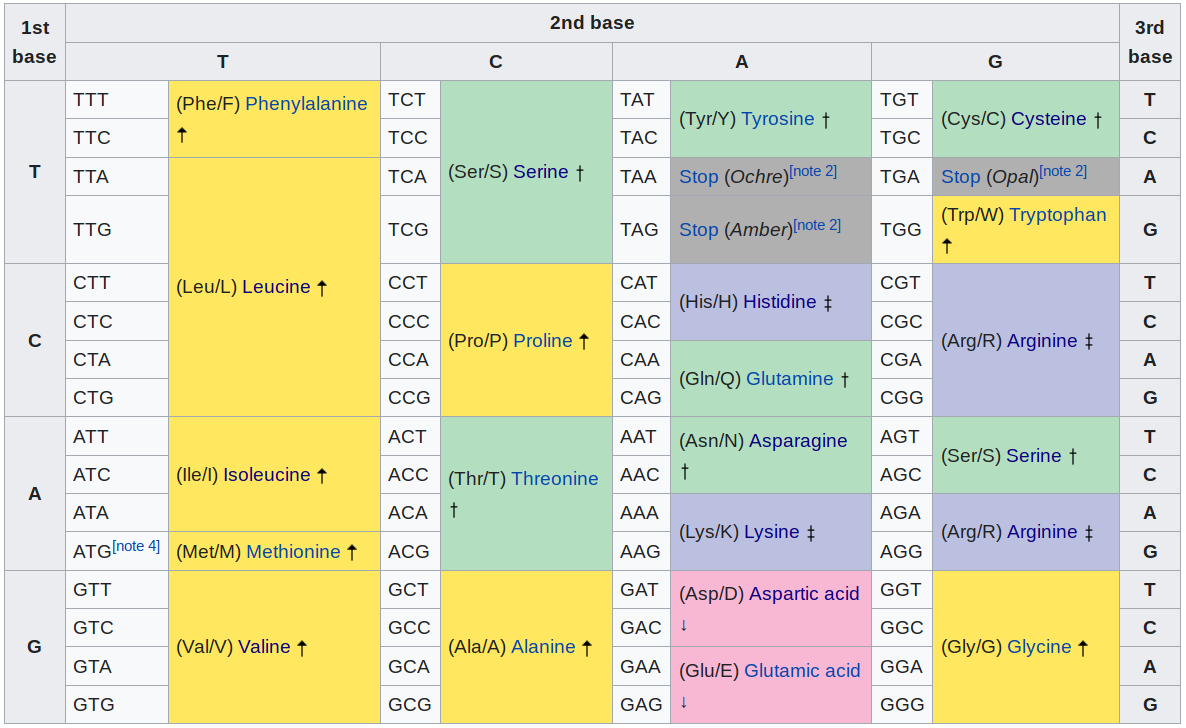
Also, the codon translation table might come handy
Example
- hg 19 chrX 12906010 G>T; PDB identifier: 6lw1
Exercises
- Today's special - use the "insert game to illustrate in a slide how an early stop codon happens.
- Then, in each exercise use the UCSC genome browser to answer the question: "What is the consequence of the variant given in the title?" (Insertion or deletion, missense mutation or an early stop codon, splice site change ...)
- If the structure is known ( the PDB identifier in the title), show the location of the change on the structure.
- What is known about the mutations in this gene? Consult the OMIM database.
- Does this variant exist in the general population? Consult the GnomAD database. Ho common is the variant? How common are the homozygotes (what are homozygotes) in this variant?
- hg19: chr1 45798118 G>A; PDB identifier: 3N5N
- hg19: chr17 41223242 G>C
- GRCh37:ChrX:49107935 G>A; PDB identifier: 4wk8
- GRCh37: Chr14:74947484 T>C
- GRCh37: Chr3:33156041 G>C
- GRCh37: Chr17:41833024 C>T; PDB identifier: 6l6r
- GRCh38 chr15:90883330 G>A
- why is this variant interesting? Read the abstract here.